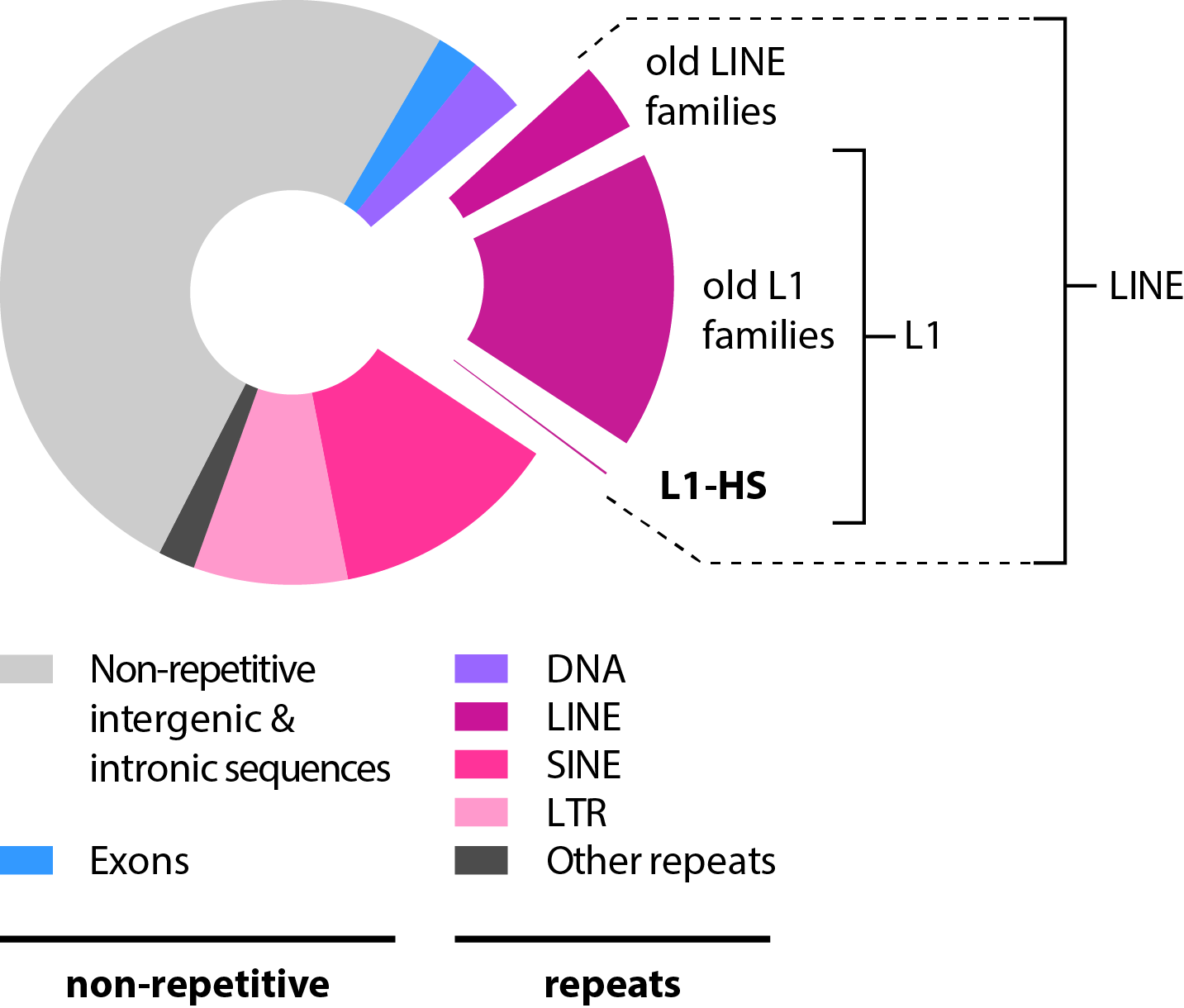
| Retrotransposons, which comprises LINE, SINE and LTR-containing elements, accounts for almost half of our genome (Fig. 1). They are mobile genetics elements - also known as jumping genes - but only the L1-HS subfamily has retained the ability to jump autonomously in modern humans. Their mobilization in germline - but also some somatic tissues - contributes to human genetic diversity and to diseases, such as cancer. Additional background on L1 elements can be found here. |
 |
|
| euL1db provides a curated and comprehensive summary of L1-HS insertion polymorphisms identified in healthy or pathological human samples and published in peer-reviewed journals. It will help understanding the link between L1 retrotransposon insertion polymorphisms and phenotype or disease. |
||
| There are many ways to interrogate euL1db. You could start with these examples of euL1db use, or by reading the help or FAQ sections. If you are eager to try euL1db, why not to start by finding if your favorite gene contains known L1 insertion polymorphisms, by clicking here? |
Figure 1 - Proportion of L1-HS elements in the human reference genome. L1-HS represents ~3.3 Mb of the reference genome (~0.1%), but each individual has additional L1-HS copies not present in the reference genome (non-reference L1 copies), which contribute to our genetic diversity. On the average, two human individual genomes differ at 285 sites with respect to L1 insertion presence or absence (Ewing et al. 2011). |




