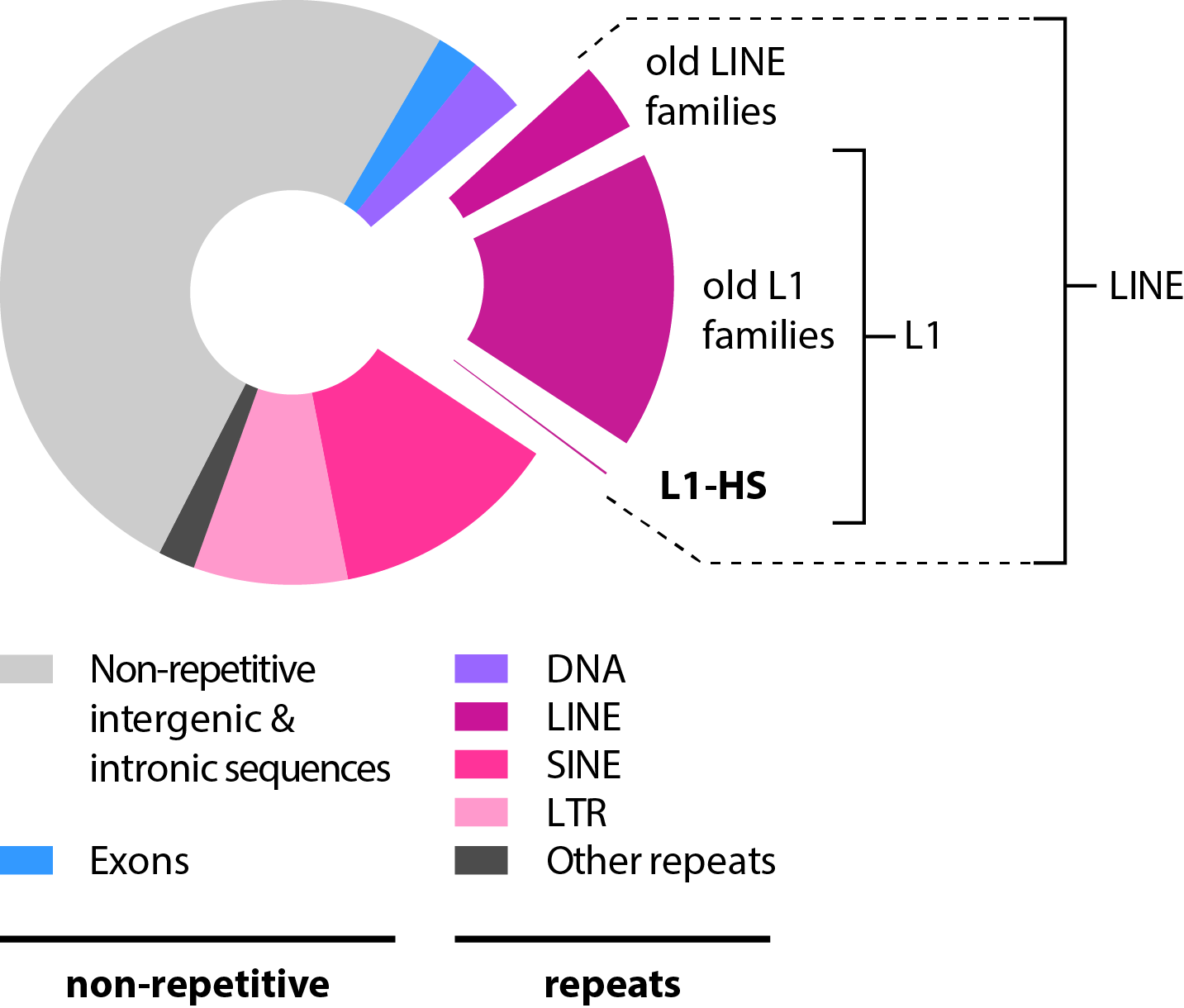
|
A full-length human L1 is ∼6.0 kb in length, contains an internal promoter located in the 5'-untranslated region (UTR) and two non-overlapping open-reading frames (ORF1 and ORF2), separated by a short inter-ORF spacer . Both ORFs are required for L1 retrotransposition . ORF1 encodes a 40 kDa protein (ORF1p) that contains a coiled-coiled domain (CC), a non-canonical RNA recognition motif (RRM) domain and a basic C-terminal domain (CTD). Figure 2 depicts the L1 life-cycle. L1 replication starts by the transcription of a bicistronic mRNA (A). The L1 RNA is exported to the cytoplasm (B). ORF1p and ORF2p proteins are translated and bind to the L1 RNA to form L1 ribonucleoprotein particles (RNP) (C). The L1 RNP is imported into the nucleus (D). Integration and reverse transcription occur at the genomic target site. First, the L1 endonuclease (EN) activity nicks the target DNA (red arrowhead, E). Then, the L1 reverse transcriptase (RT) initiates the reverse transcription of L1 RNA through annealing between the target site and the poly(A) tail of the L1 RNA (black arrowhead, F). The mechanisms involved in the final steps of this process and the resolution of the integration are unresolved yet (G). Partial reverse transcription can lead to 5'-truncated L1 copies.
|
|
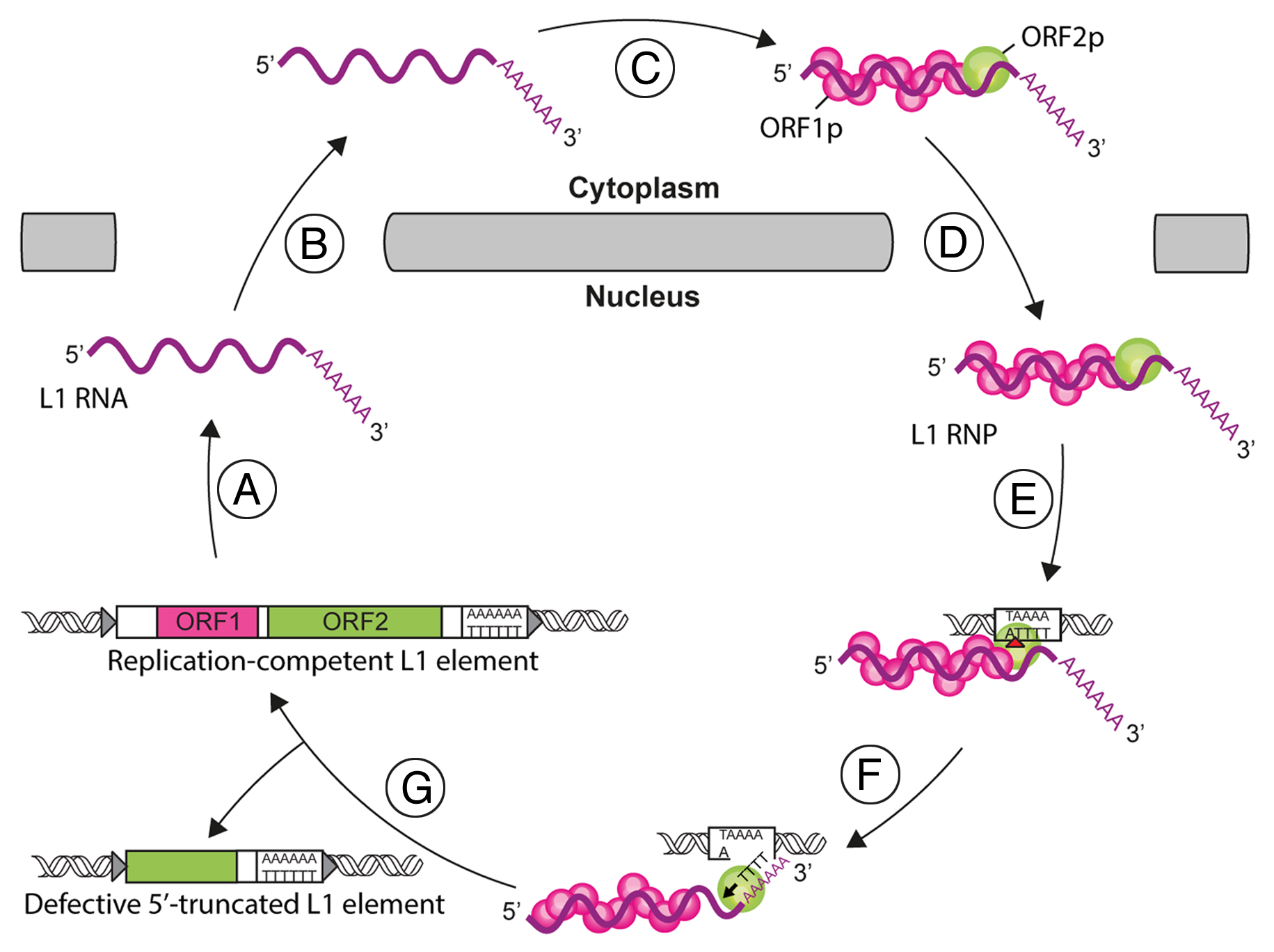
Figure 2 - L1 life-cycle. See text for details.
From Viollet et al. Mob Genet Elements 2014, License CCBY-NC [Pubmed]
|