| euL1db |
| The European database of L1-HS retrotransposon insertions in humans |
| Home | Background | Search | Browse | Utilities | Help | FAQ | Data | Contact |
|---|
| 1 - I am working on liver cancer. How do I get all L1-HS insertions, which are present in samples from the liver of patients with hepatocarcinoma? |
| 4 - I have done a genome-wide association study (GWAS) to find genetic determinants of a specific human trait. How could I test if a polymorphic L1-HS insertion might be associated to this trait? |
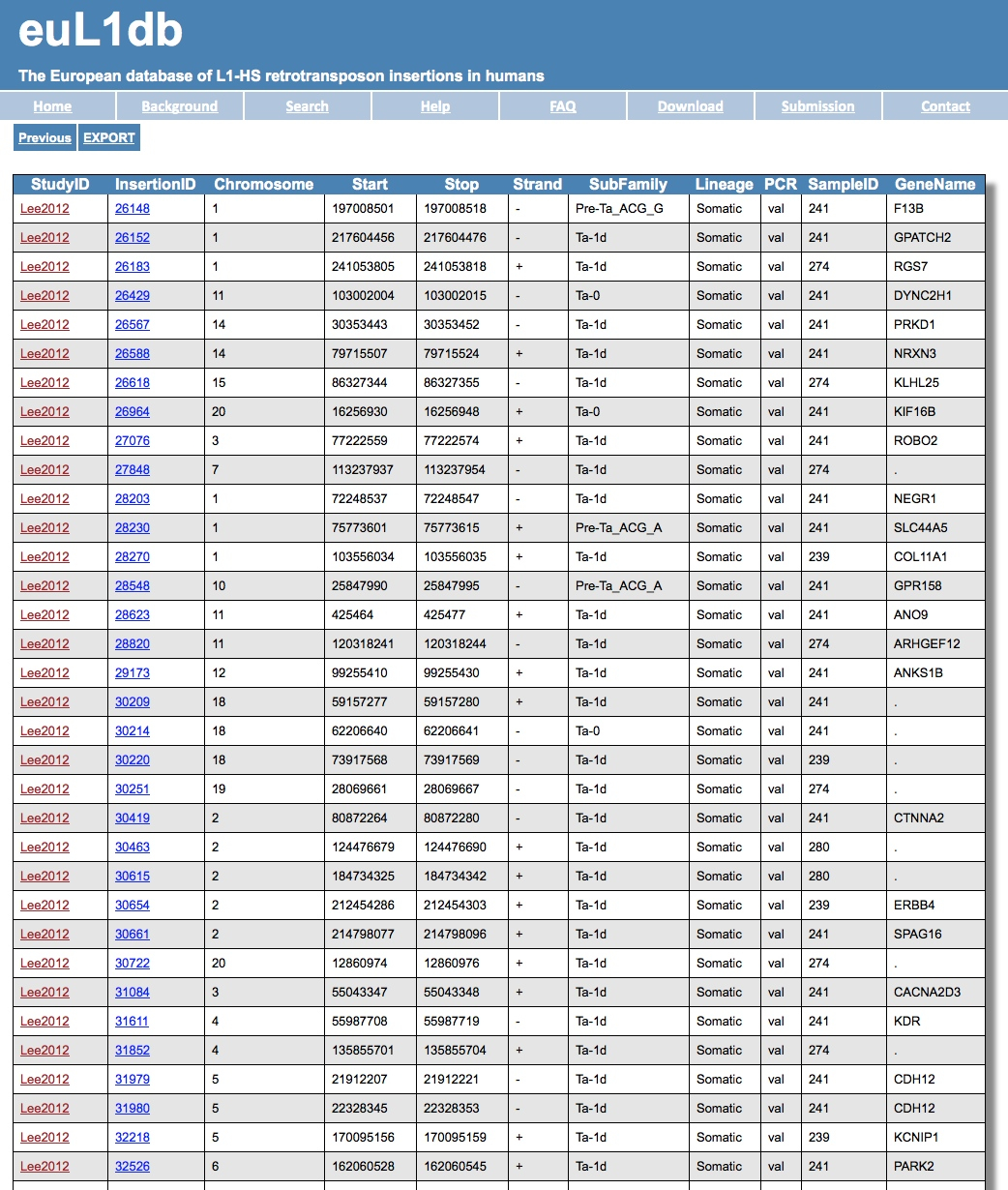
| 1 - I am working on liver cancer. How do I get all L1-HS insertions, which are present in samples from the liver of patients with hepatocarcinoma? |
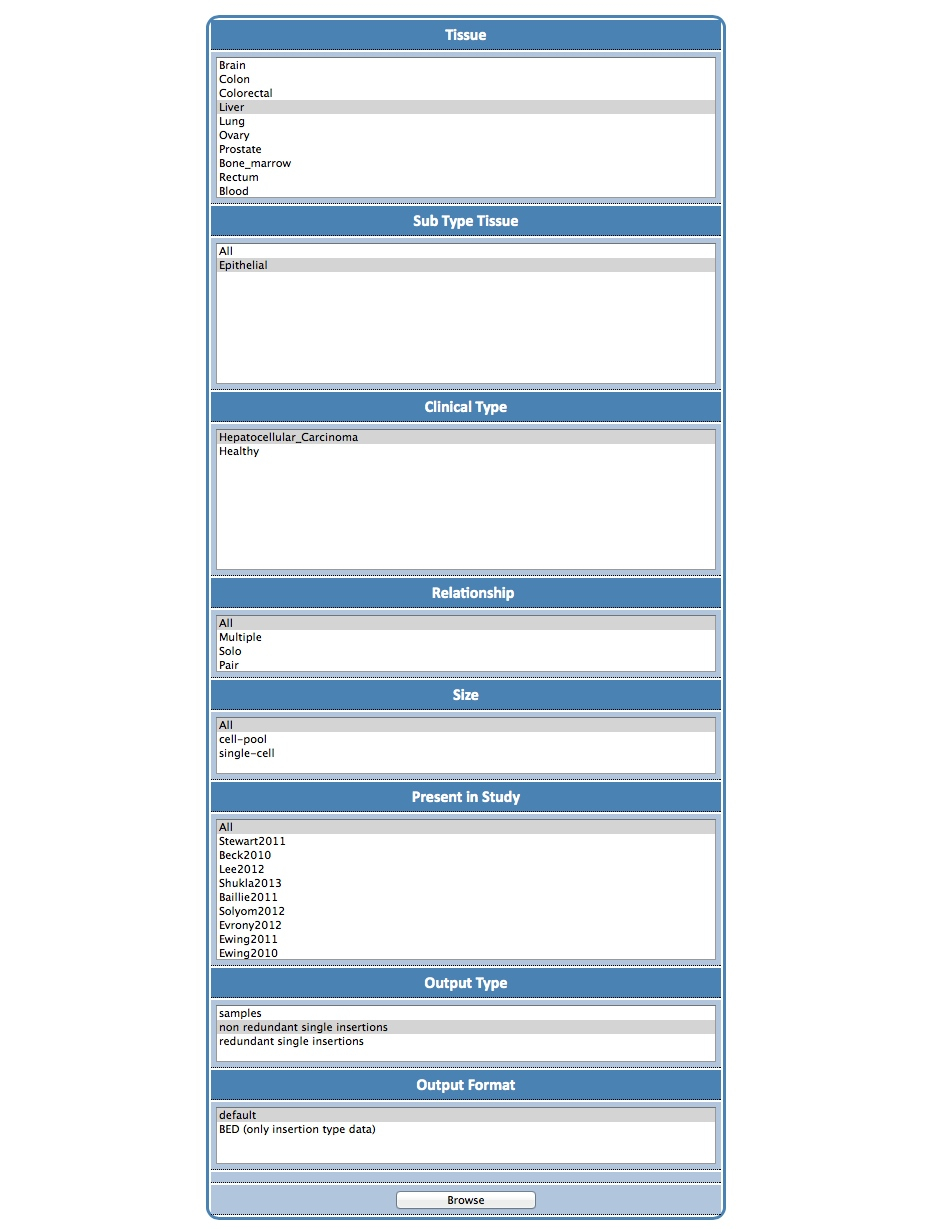
| The steps are: (1) in the Search tab, go to the Samples Browser; (2) select Liver as a tissue; (3) then Epithelial/All as a sub-tissue type; (4) then select Hepatocellular Carcinoma as clinical type; (5) finally, choose your output type (e.g. non-redundant insertions). |

| The result is a table with a list of insertions. You can then export the results into a file using the EXPORT option on top left hand side. Like the page below: |
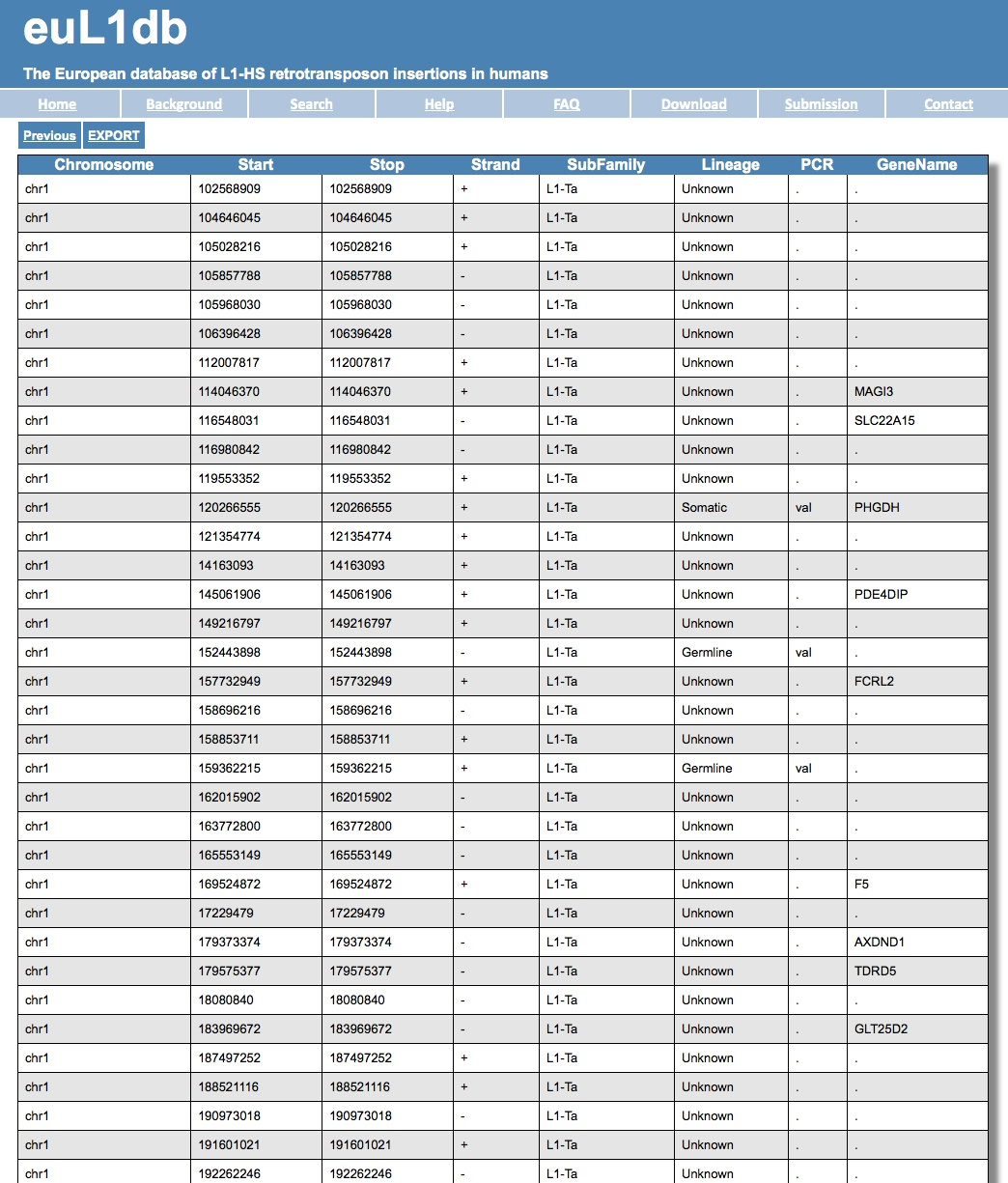
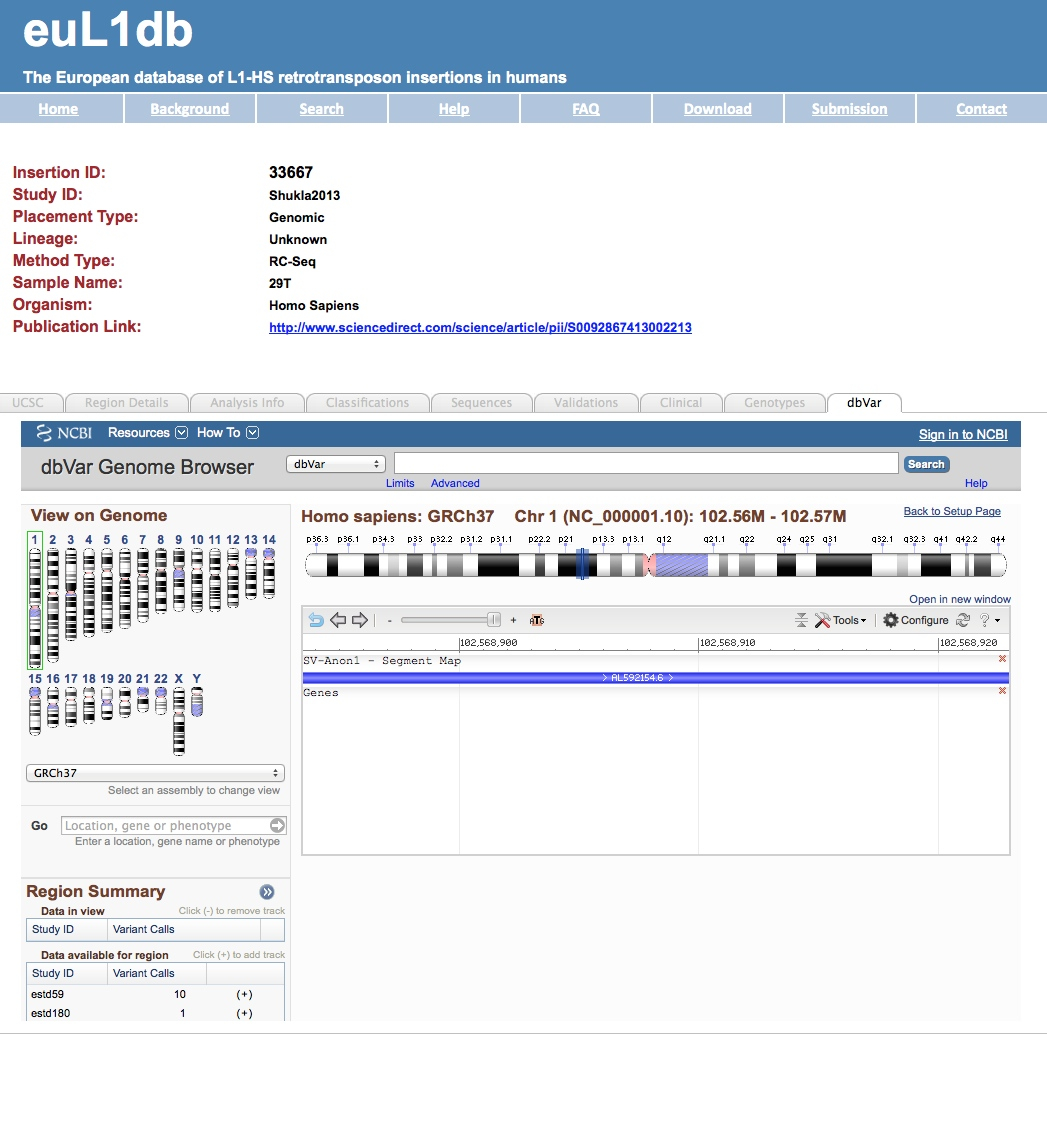
| If you had selected as Output type the option “redundant single insertions”, you can also get for each insertion a detailed page with annotations including dynamic links to the UCSC or dbVar genome browser for every insertion. |
| The steps are: (1) in the Browse tab, go to the Insertions Browser; (2) in the Lineage drop down menu select "Somatic"; (3) finally, press the Browse button and here you go ! |
| (1) in the Browse tab, go to the Insertions Browser. |

| (2) in the Lineage drop down menu select "Somatic". |
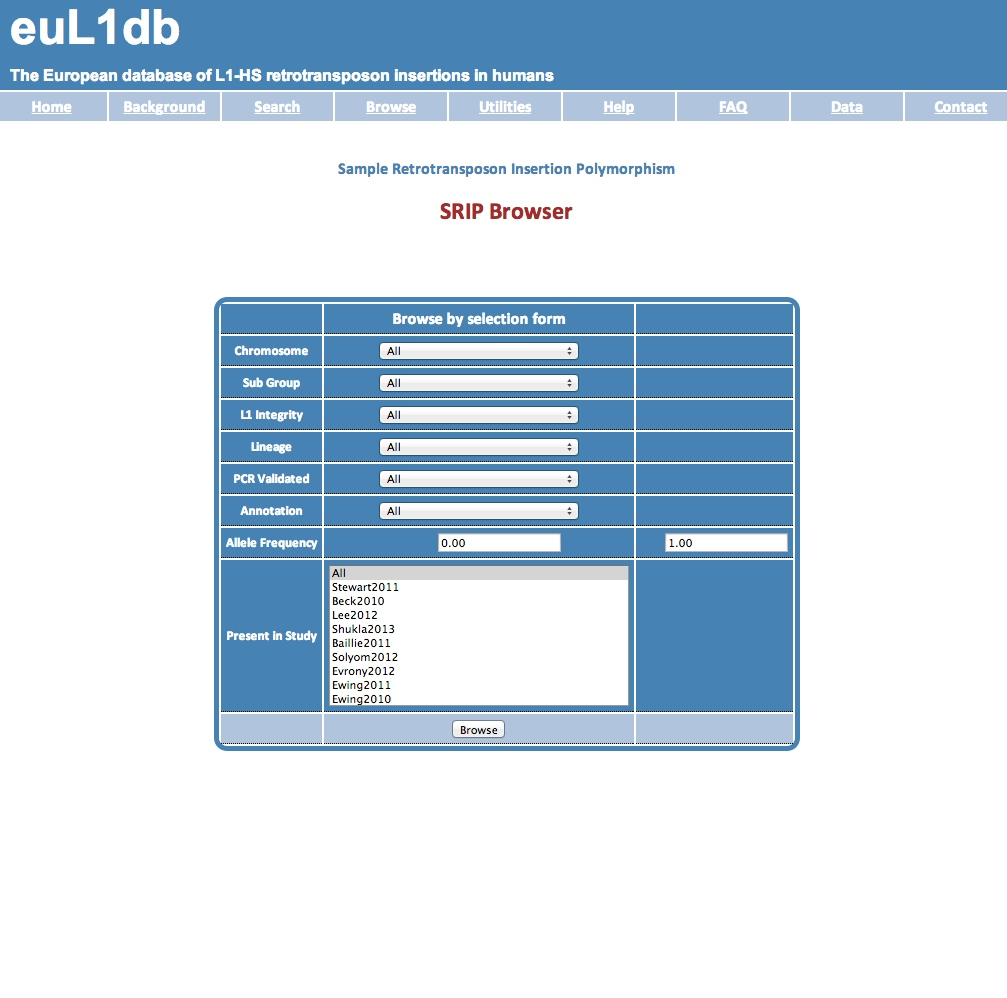
| (3) Finally, press the Browse button and here you go ! You will get a list of SRIP. For those falling in genes, the gene name is provided as last column. You can then export the results with the Export button to retrieve the gene list. Then you can use this gene list in your favorite Gene Ontology enrichment software (e.g. DAVID, GOrilla, AmiGO, EasyGO, Enrichr, GREAT). |
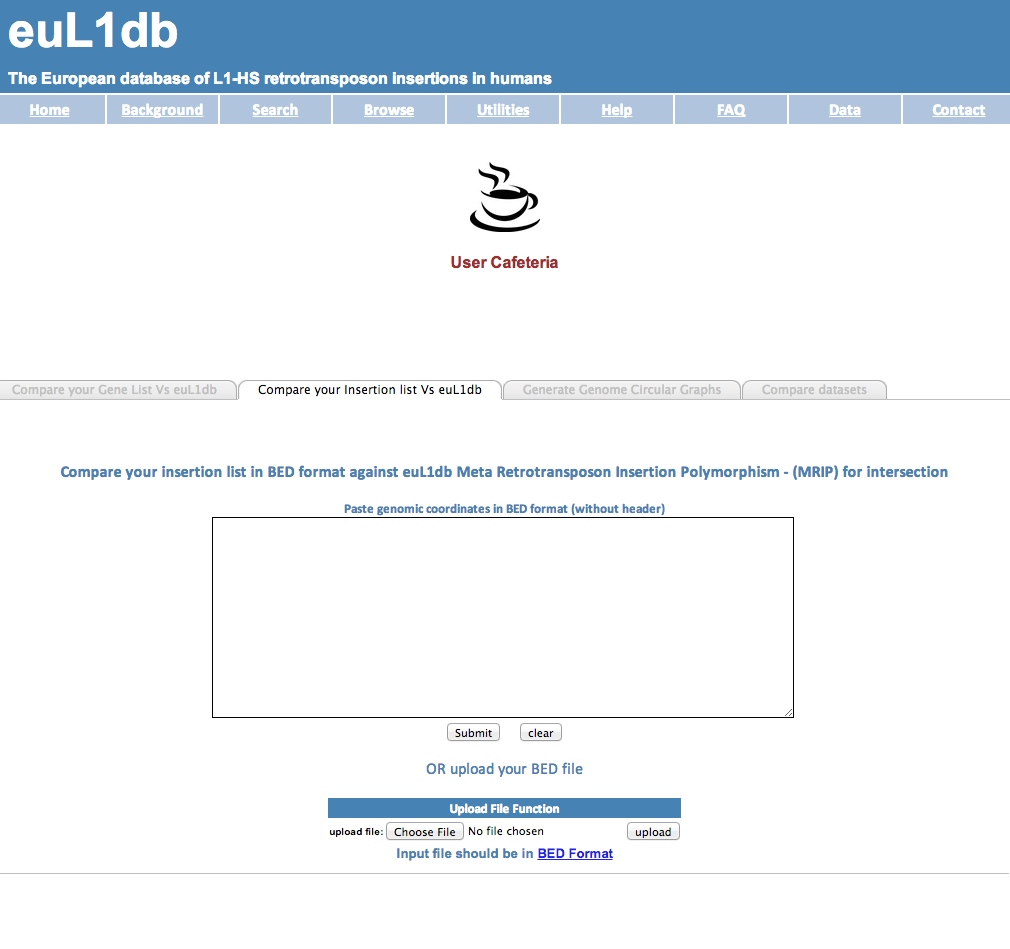
| You will need a text file with the genomic coordinates of your candidate L1-HS insertions (in BED format). The steps are: (1) in the Search tab, go to the Utilities Browser; (2) click on the "Compare your insertion list Vs euL1db TAB"; (3) then upload your file and here you go! |
| (1) in the navigation panel click on the Utilities Browser |
| (2) Click on the "Compare your insertion list Vs euL1db TAB". (3) Upload your file and here you go ! |
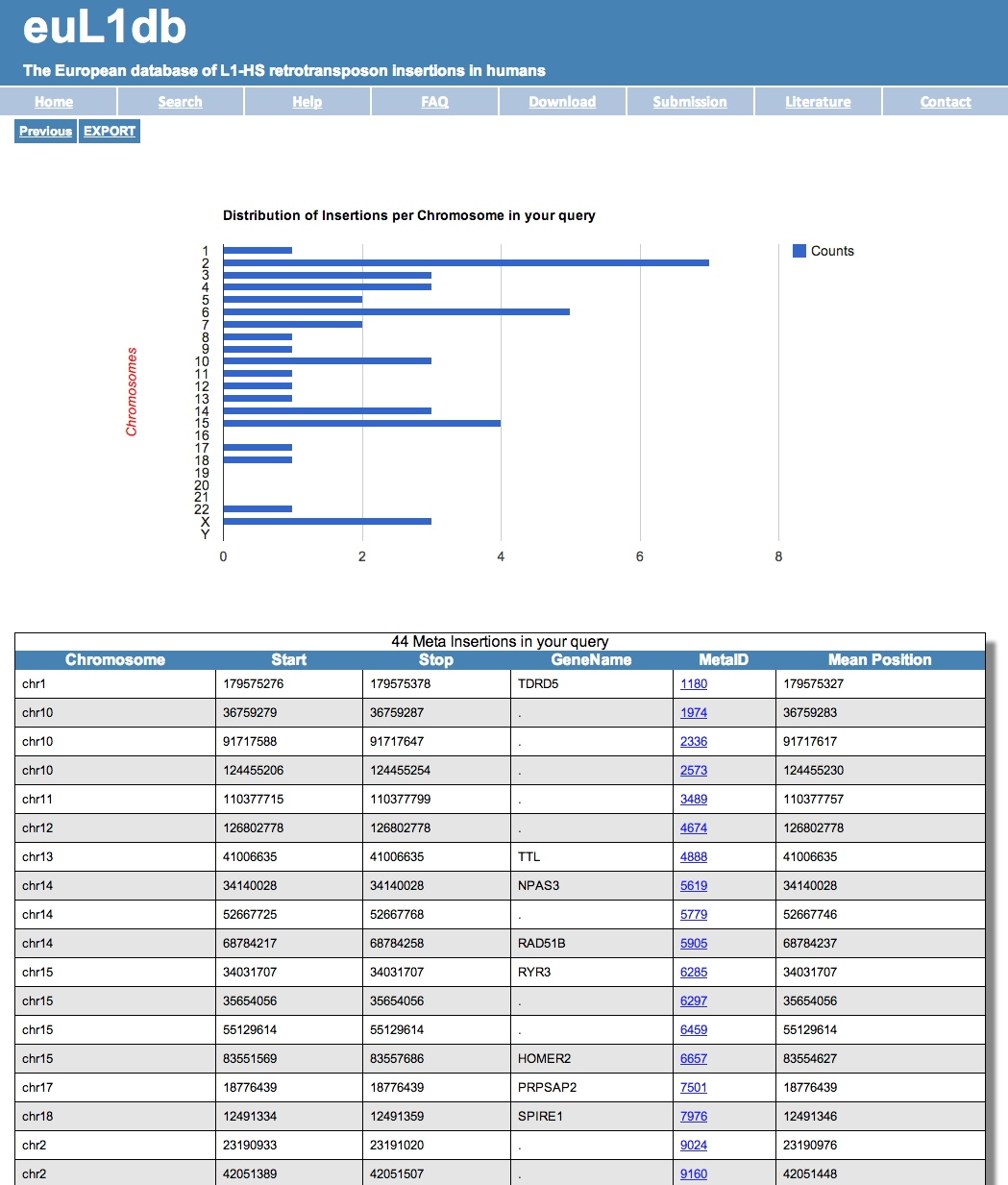
| You will get a list of MRIP as a result page like below. You can also use the links provided in the output page to zoom down to SRIP. You can then export the results and save them. |
| 4 - I have done a genome-wide association study (GWAS) to find genetic determinants of a specific human trait. How could I test if a polymorphic L1-HS insertion might be associated to this trait? |
| The steps are similar to the example 3. You will need to upload a list of genomic coordinates corresponding to your candidate SNPs instead of the L1-HS coordinates. |