| euL1db |
| The European database of L1-HS retrotransposon insertions in humans |
| Home | Background | Search | Browse | Utilities | Help | FAQ | Data | Contact |
|---|
| euL1db is a database of human-specific L1 (L1-HS) insertions and allows its user to search, view, and download known published L1-HS insertions. euL1db is a free resource which is developed and maintained by the Institute for Research on Cancer & Ageing of Nice (IRCAN), France. |
| Concrete examples on how to use euL1db to answer specific biological questions are displayed here. |
| Data structure of euL1db can be broadly summarized in 6 data types which are interconnected in a dynamic way. Below is a brief explanation about each of them: |
| (i) Studies: All insertion regions and insertion instances, individuals and samples that are submitted as a group are part of a study. Each study typically represents a coherent set of methods and analyses that were performed at around the same time, by the same authors, in the same laboratory (or laboratories). Because these parameters determine to a large extent the variability that exists between datasets, all data in euL1db are organized by study. Typically, a study will correspond to a single publication or community resource. |
| (ii) Individuals: All individuals that are submitted as a part of a study refer to the source individuals from whom the data samples were originally taken from. There are generally more than one individual for a given study. The same individual might have been subjected to multiple analyses, eventually in different studies. When available, euL1db stores the gender, geographical and family information. Each individual is given a unique ID. |
| (iii) Samples: All samples that are submitted as a part of a study refer to the samples taken from a given individual. When available, euL1db stores the clinical, Tissue, Sub-tissue, disease state and the individual from which it was taken. Each sample is given a unique ID. |
| (iv) Families: Family refers to any kind of familial relationship between two or multiple individuals within or across studies. Each family specifies a group of individuals which could have different clinical conditions and could vary in sex and age. When available, euL1db specifies the kind of relationship between individuals and assigns a unique ID to the family. This can help in genetic studies.If an individual has no known connection with other individuals in euL1db, then it is described as 'solo'. |
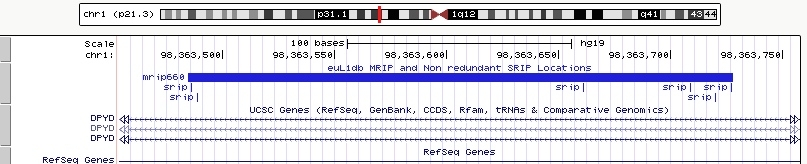
| (v) Sample Retrotransposon Insertion Polymorphism - (SRIP): An SRIP is a location within the genome that a submitter has defined as containing an L1-HS insertion in a given sample. An SRIP is typically defined by its genomic coordinates and information like Sequence, Peak ranges, Insertion break point information and, when available, other informations like genomic strand, Target Site Duplication length and sequence, 3' prime transduction, polyA size, position on the reference L1HS, allele frequency, if it is a somatic or germline insertion, and integrity (full length or truncated L1-HS). Each SRIP is given a unique ID in euL1db, which is prefixed by srip (e.g. srip34564) and can have duplicate genomic coordinates with other insertions from different samples. |
| (vi) Meta Retrotransposon Insertion Polymorphism - (MRIP): An MRIP refers to a unique range within the genome which contains overlapping and intersecting SRIP. Different methods and studies have variable accuracy in defining the precise location of L1-HS insertions. MRIP pools SRIPs wihin a range to remove redundancy and help comparing SRIPs within or across studies. Each MRIP is given a unique ID in euL1db, which is prefixed by mrip (e.g. mrip1234). |

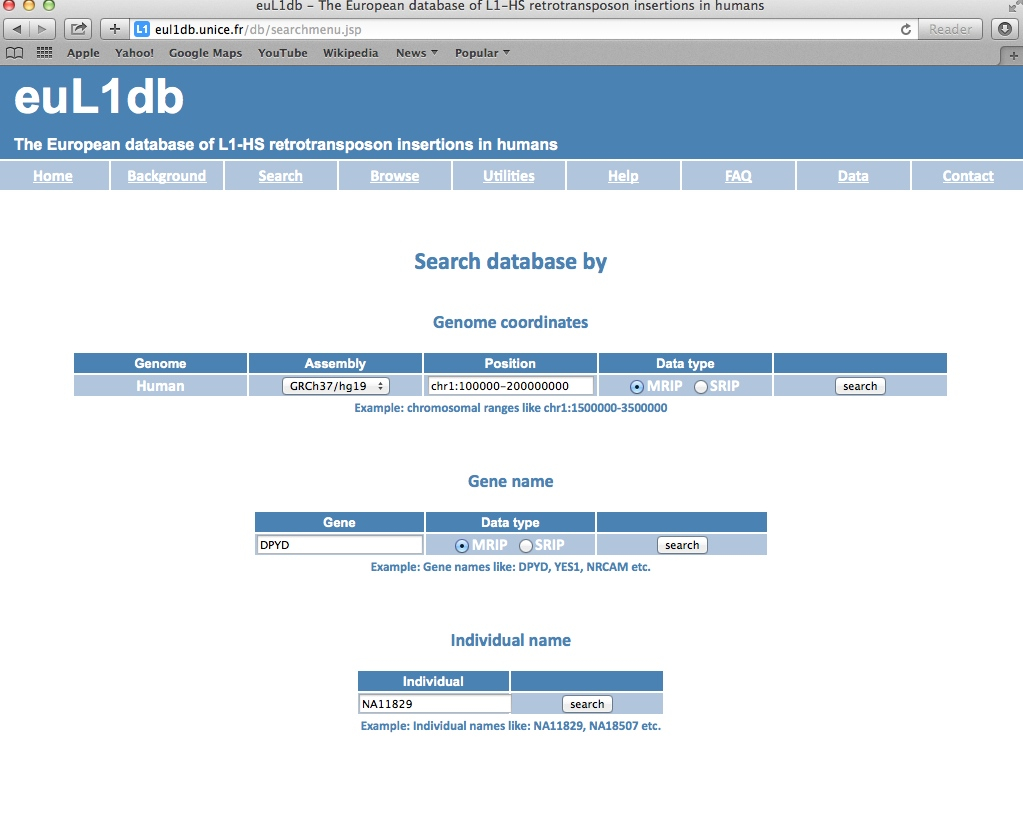
| The euL1db search fields allow extracting L1-HS insertions in a specific region of the genome, through genomic coordinates or gene name. There is also a search box for Individuals. There is an option to select the data type either SRIP or MRIP. |
| Data in euL1db is organized by Study. "Study" typically refers to a publication. It is given a study ID of the form "First Author Name" + "year of publication" and is associated with a link to the original publication. |
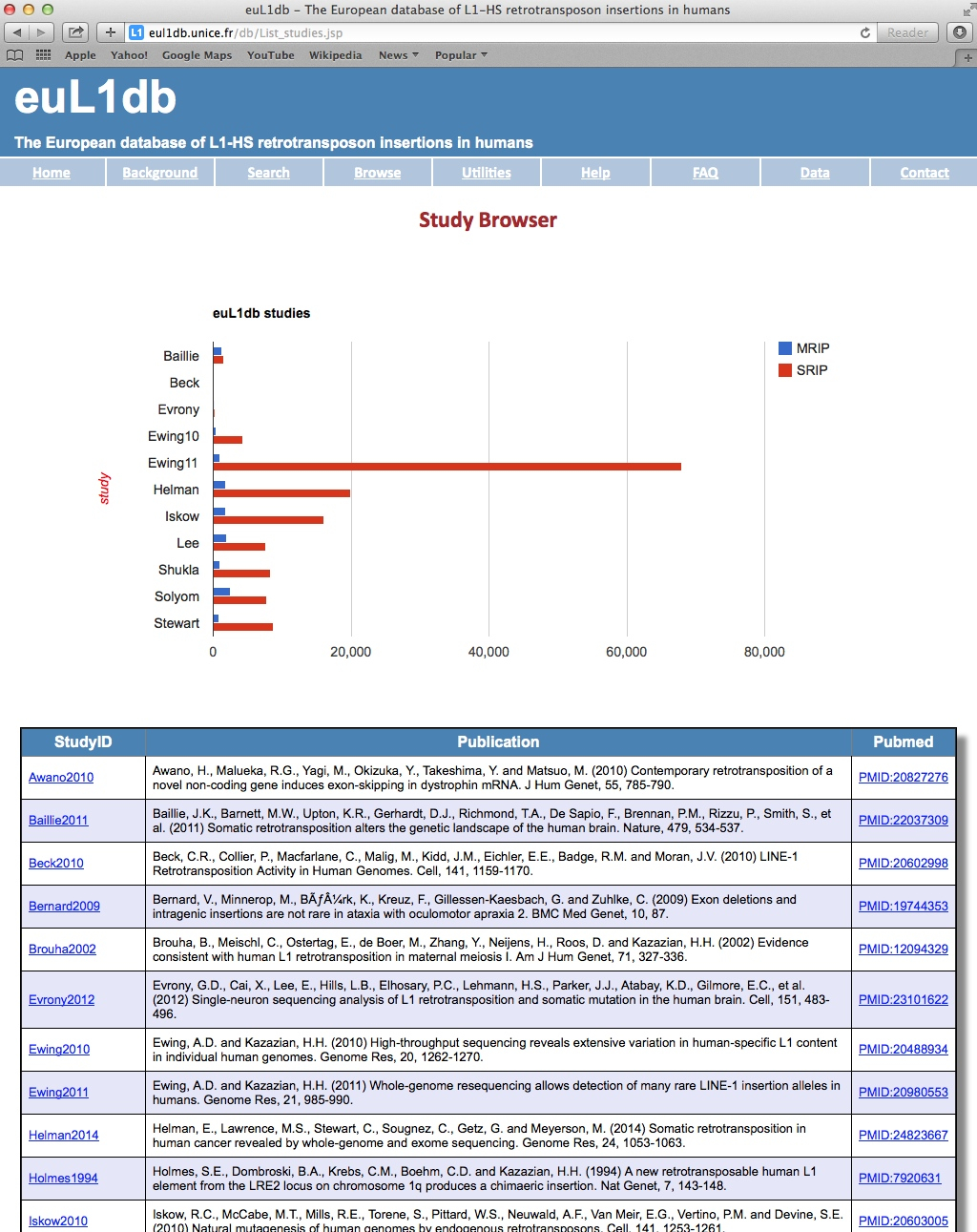
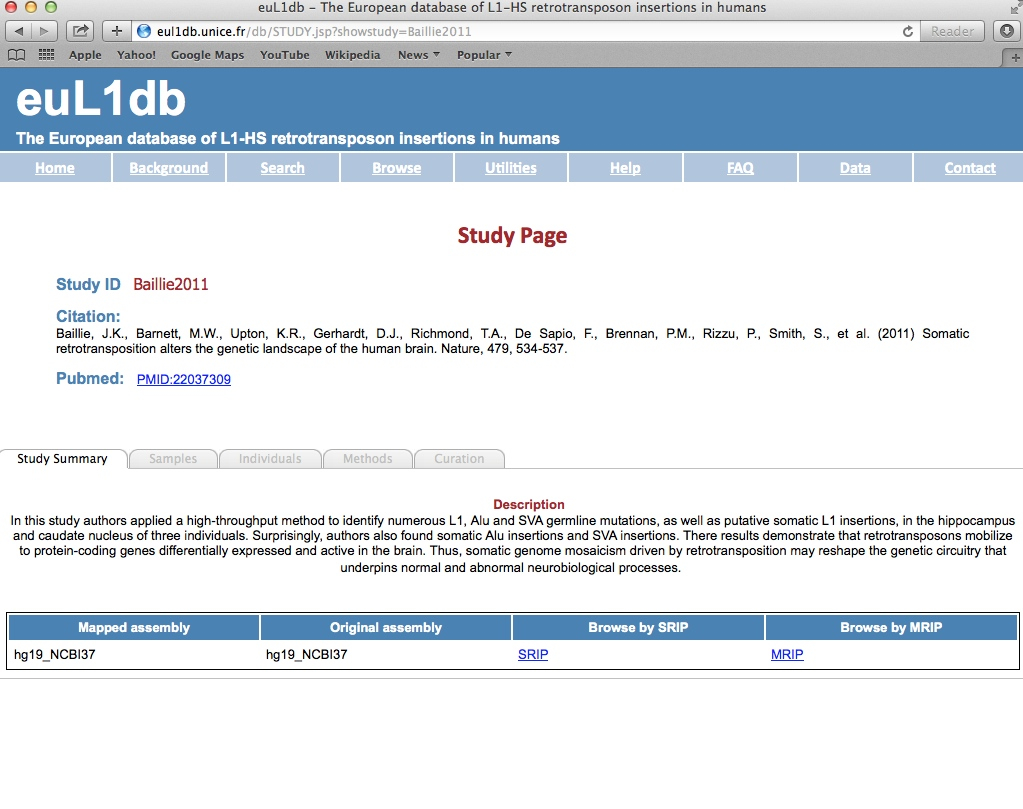
| General information: Each Study in euL1db has a Study Page that displays the general information about the study at the top, followed by a Detailed Information section organized in sub-tabs, which includes a study summary, the samples and individuals analyzed in this particular study, and a description of the methods used to find the L1-HS insertions. It also has got a tab for curation process. There is also an option to browse by SRIP or MRIP data types for the study. |
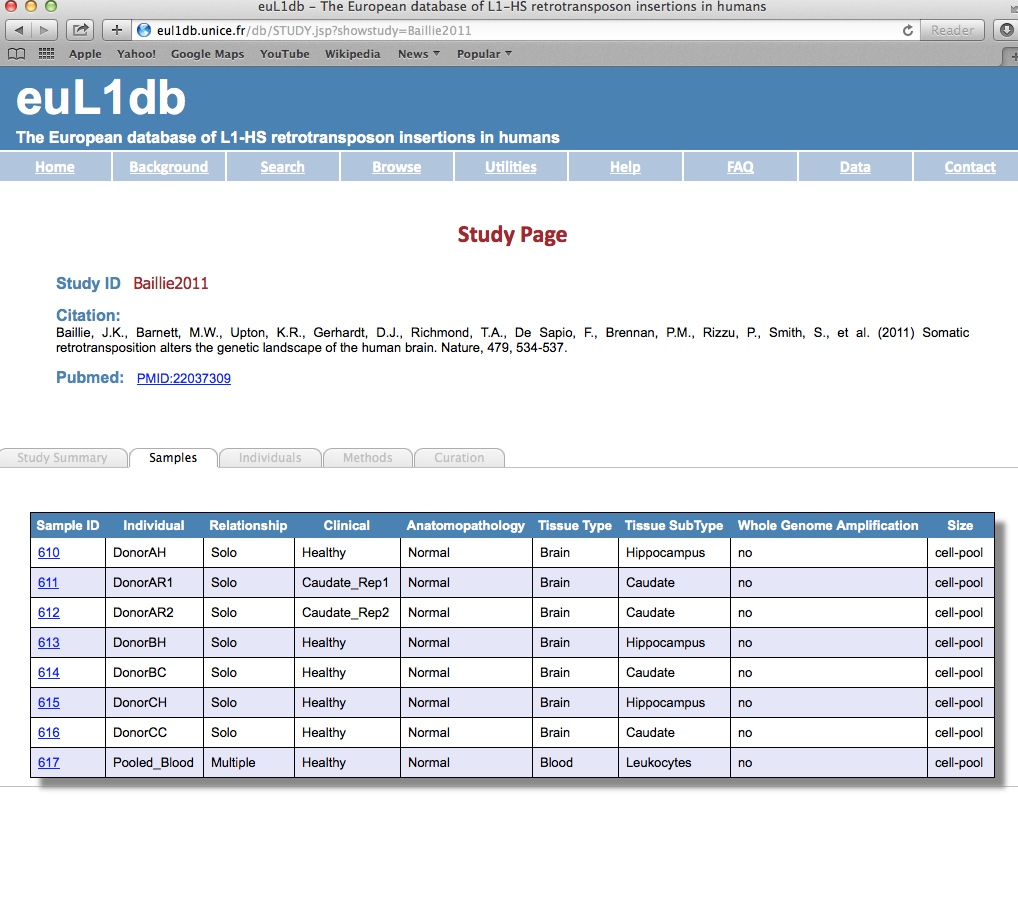
| Samples tab: The Sample tab provides details on the samples of the selected study. It includes the Sample ID, the individual ID, the potential relationship with other samples from the same individual, histological and pathological informations, whether it was obtained by whole genome amplification, and if it was a single-cell or pool-of-cell sample. Active links are provided to further interrogate and zoom down to the insertion level. |
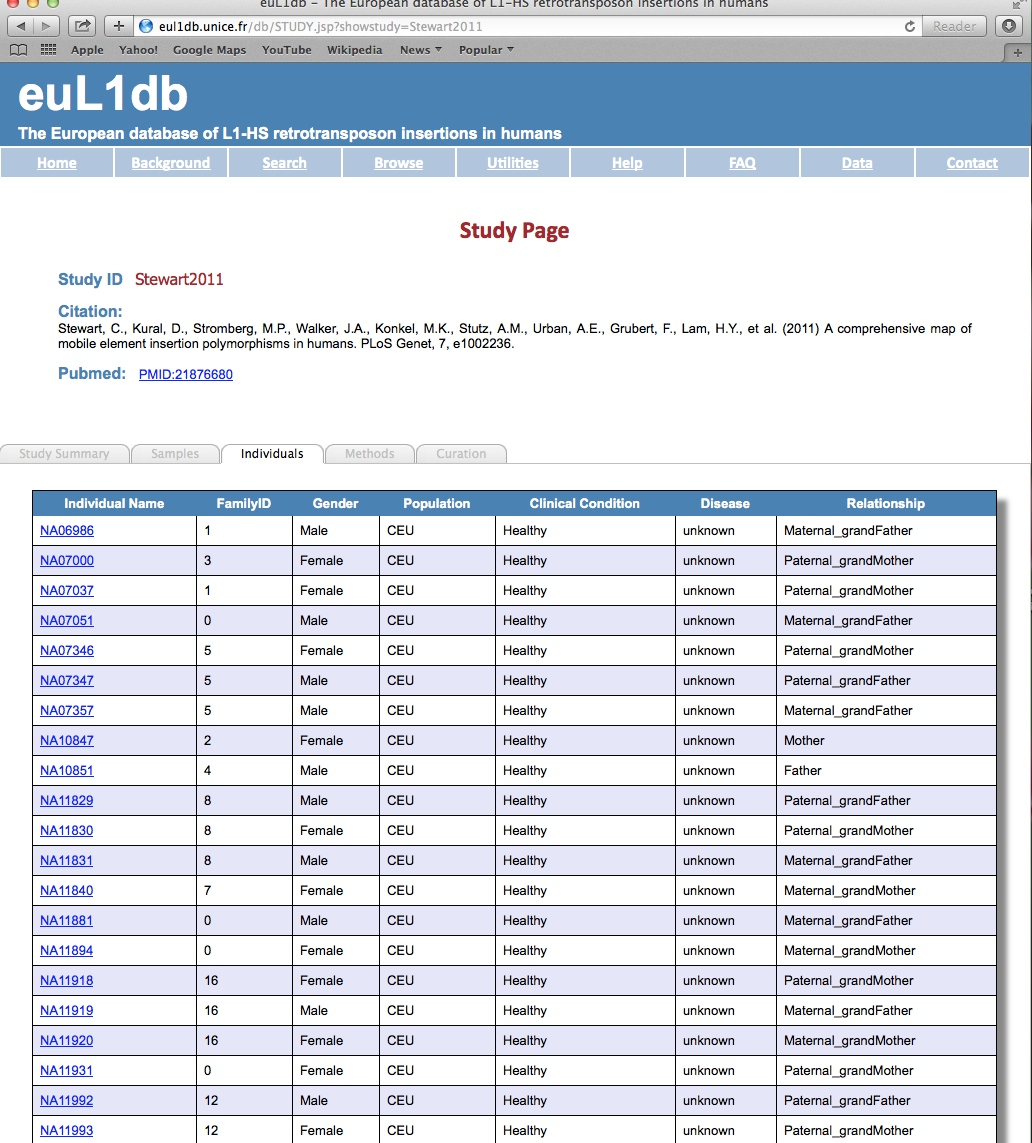
| Individuals tab: The Individuals tab provides details on the individuals of the selected study. It includes the individual name, its family ID, its gender, its geographical/ population information, its clinical condition and possible disease, and its situation in the family tree. Active links are provided to further interrogate and zoom down to the insertion level. |
| Methods tab: The Methods tab describes the methods used to find L1-HS insertions in the selected study. It includes the name of the method, a short description, a link to the reference method and the sequencing technologies used. |
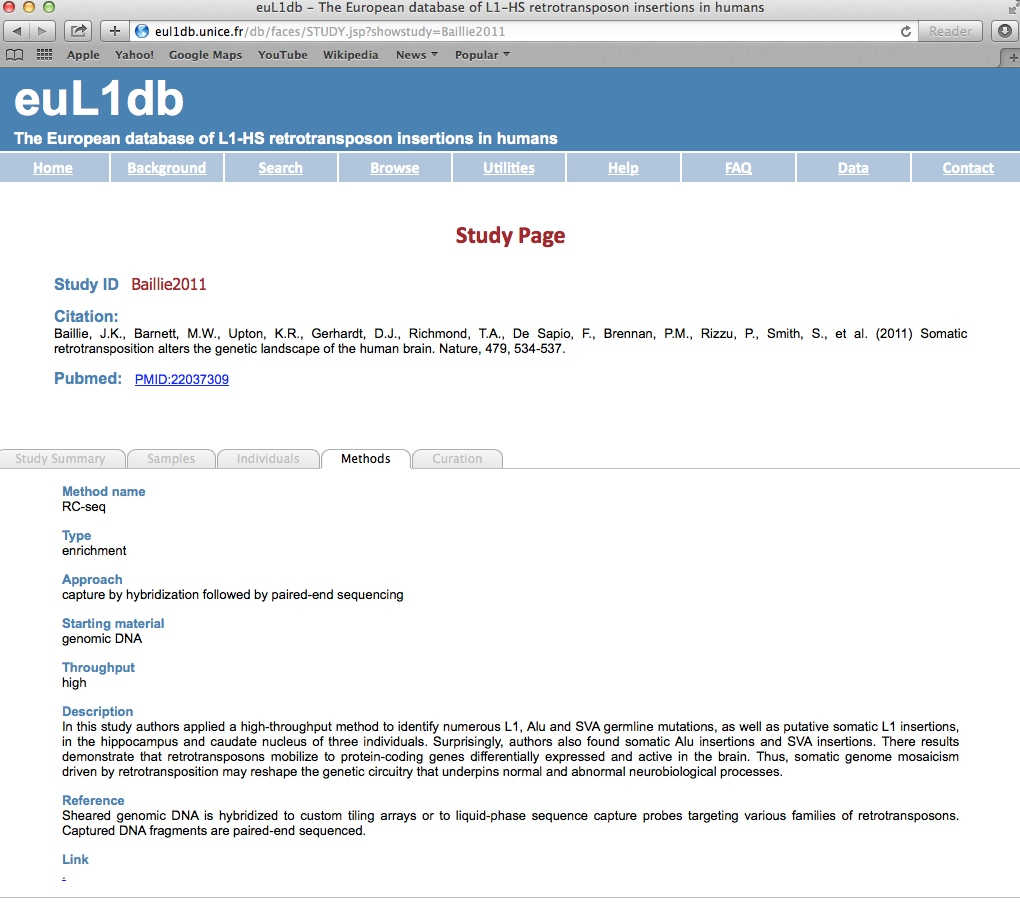
| Curation tab: The Curation tab describes the data curation source, approach and validation steps. |
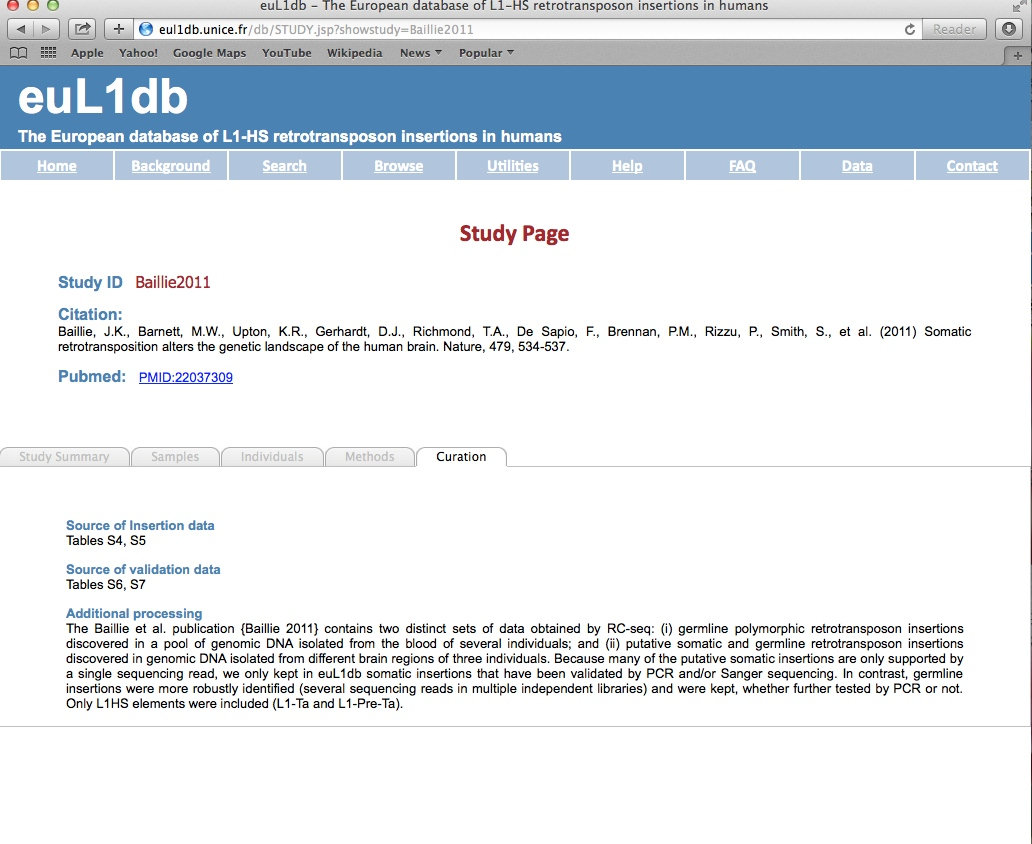
| The Individuals Browser provides a systematic way to interrogate and mine data in euL1db through individual characteristics and to retrieve their L1-HS insertions. Possible selection criteria includes: sick or healthy individuals, disease type, gender, familial relationship, or geographical/population of origin. Finally the Output Type and Format options allow the user to select the desired result table (a list of individuals, of samples or of insertions), and the format of the output. |
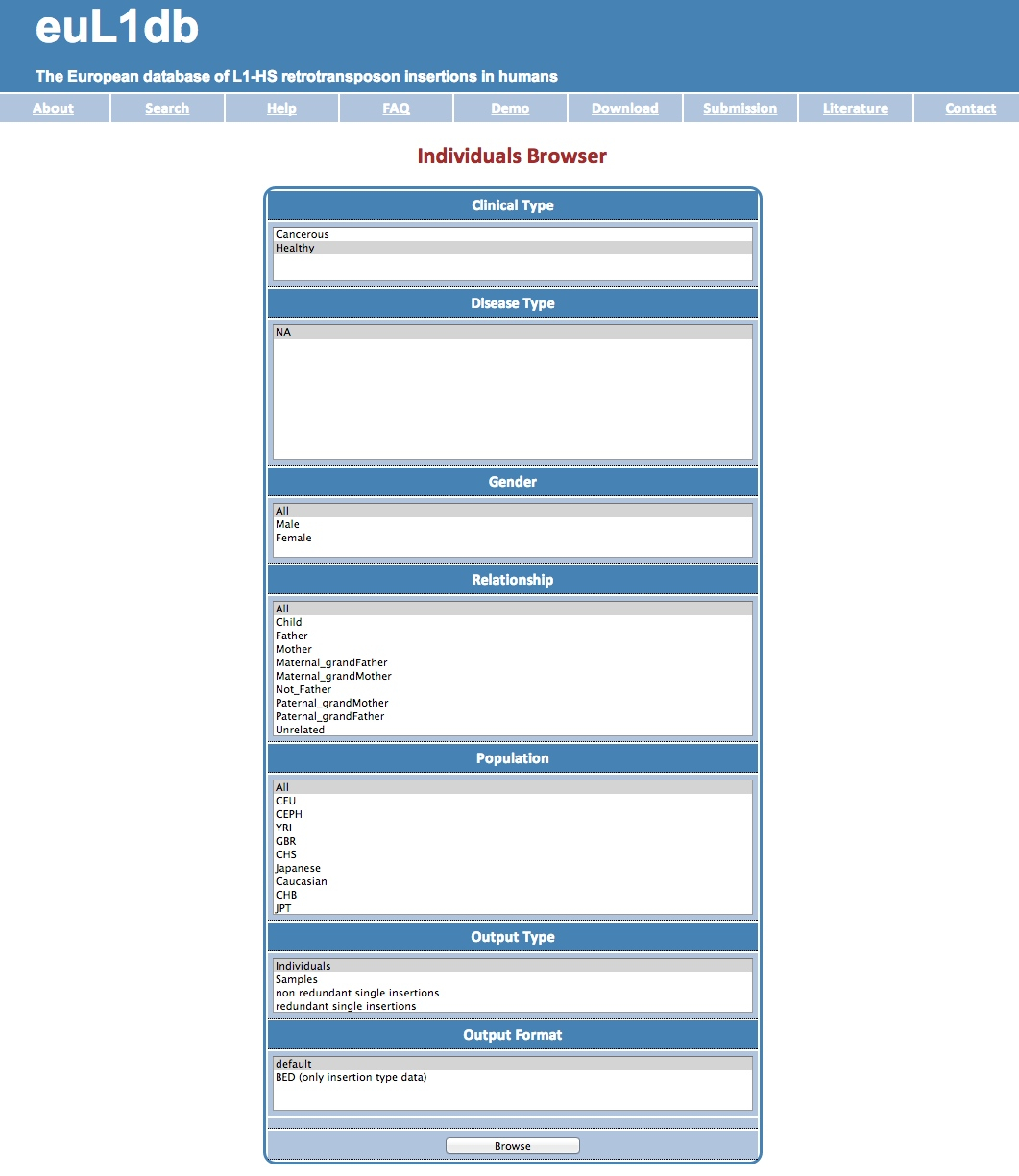
| The Samples Browser provides a systematic way to interrogate and mine data in euL1db through sample characteristics and to retrieve their L1-HS insertions. Possible selection criteria includes: tissue type and subtype in the human body, pathological state, relationship with other samples, size of the sample (single-cell or pool of cells), or the study in which it was processed. Finally the Output Type and Format options allow the user to select the desired result table (a list of samples or of insertions), and the format of the output. |
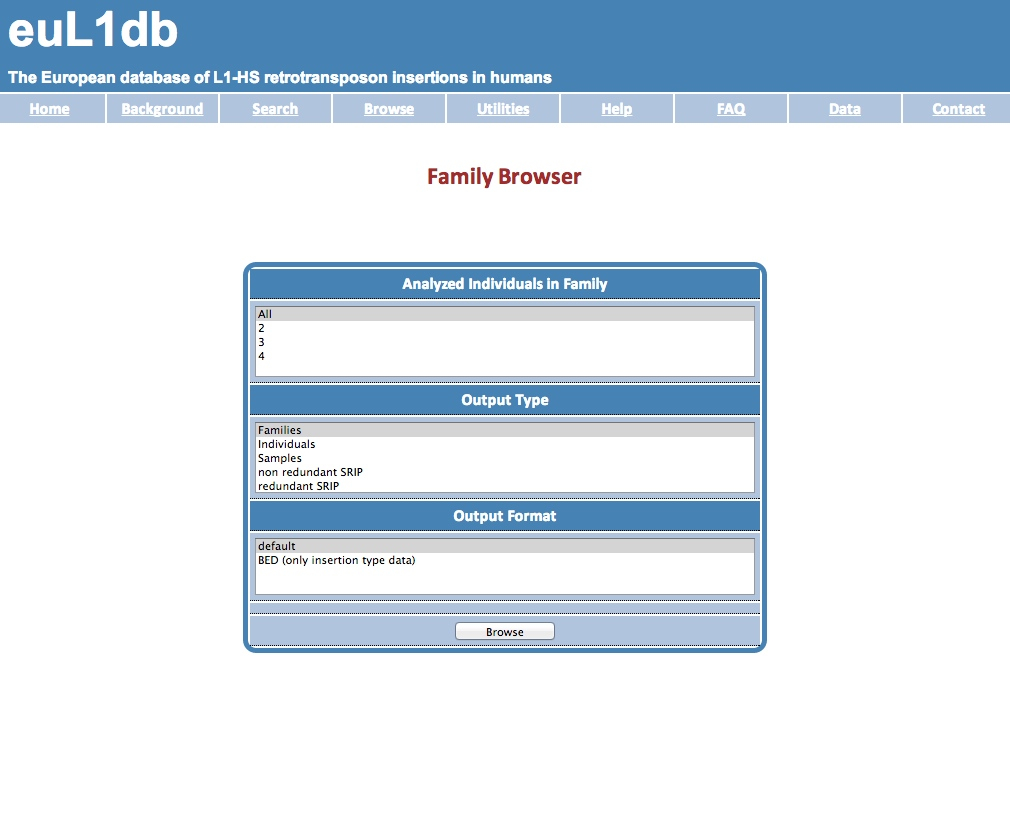
| The Families Browser allows fetching the data of related individuals. This can be useful for genetic studies. Selection is based on the members of the family analyzed. The Output Type and Format options allow the user to select the desired result table (a list of individuals, of samples or of insertions), and the format of the output. |
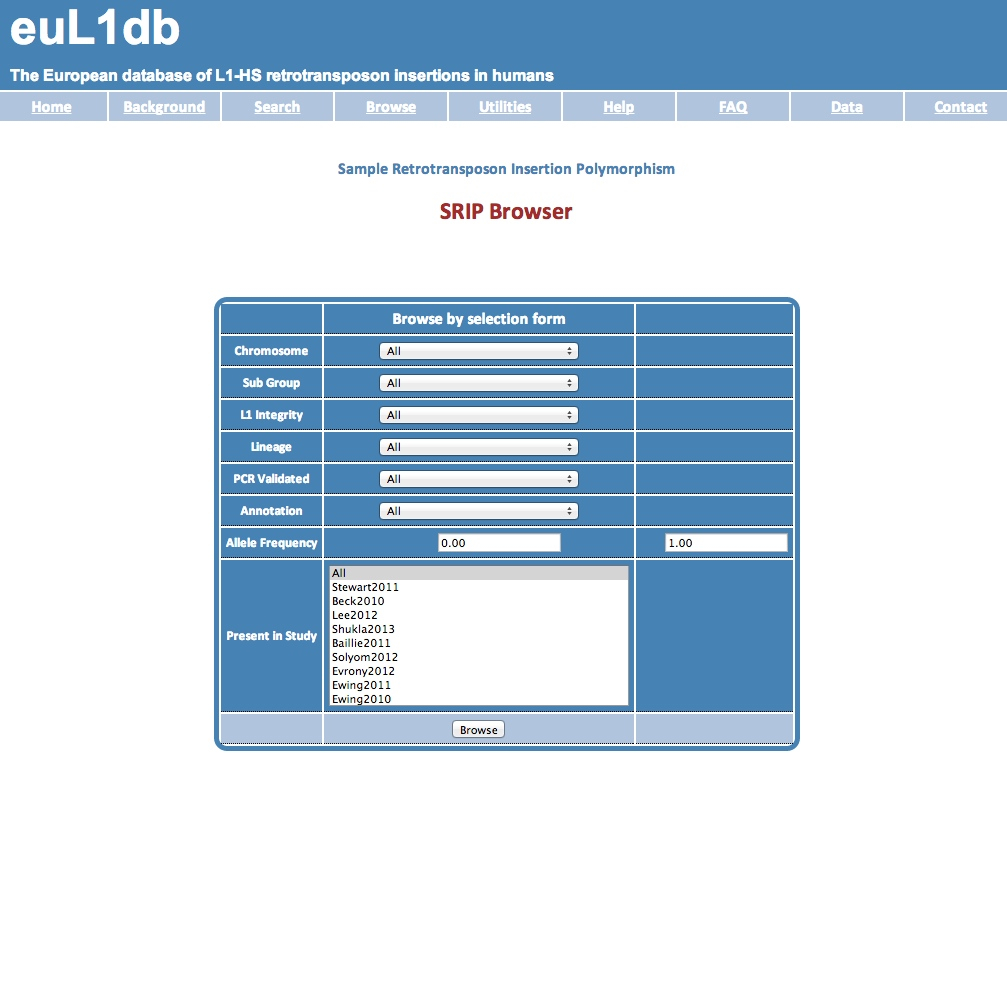
| The Insertions Browser provides a systematic way to interrogate and mine data in euL1db through insertion characteristics. Possible selection criteria includes: the L1-HS subgroup (e.g., L1-Ta or L1-Pre Ta), the integrity of the L1-HS copy (i.e. truncated or full length), the somatic or germline nature of the insertion, the level of experimental validation, the genomic location, the allele frequency, or the presence in a particular study. The output of this browser is a set of SRIP, therefore it is highly redundant. |
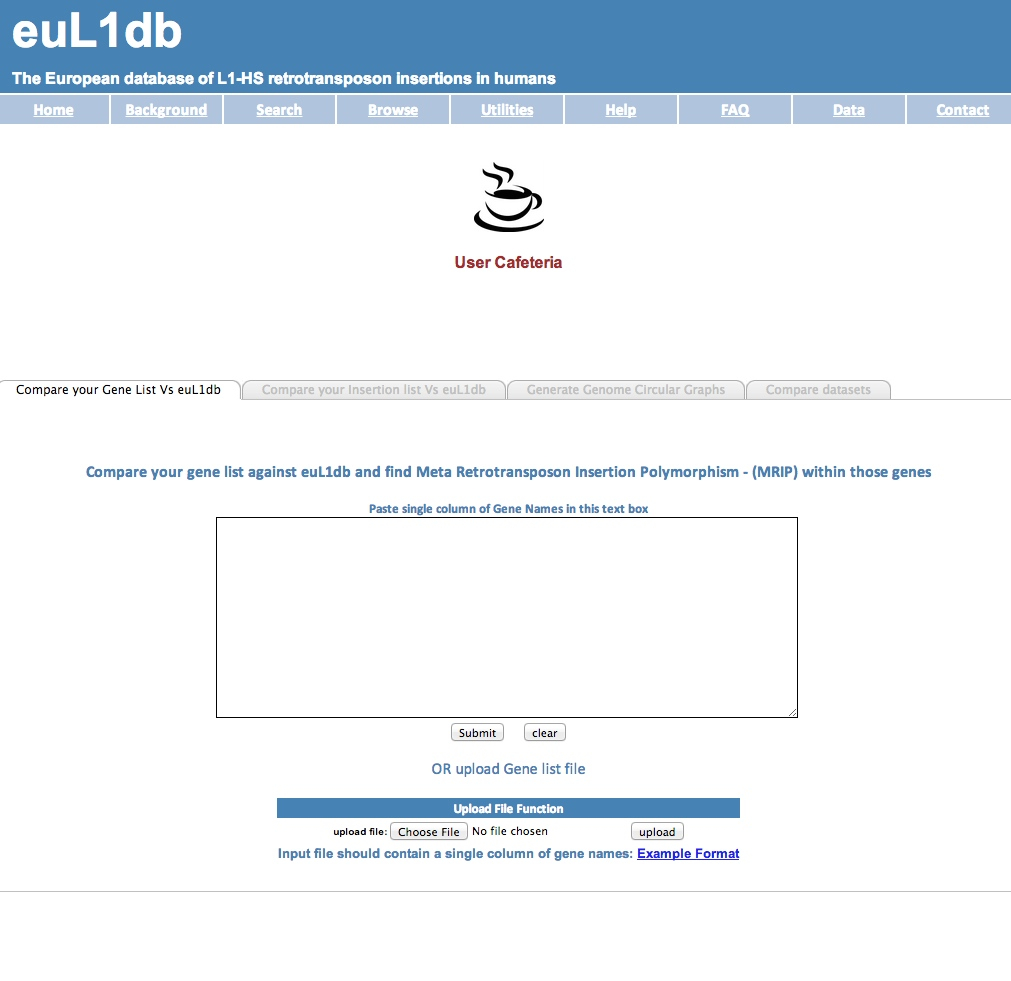
| The Utilities are a set of 4 embedded analytical tools, which allow to compare or to display samples. They are accessible from a tabbed menu and comprise the possibility to: |
| (A) retrieve the MRIP overlapping with a custom gene list; |
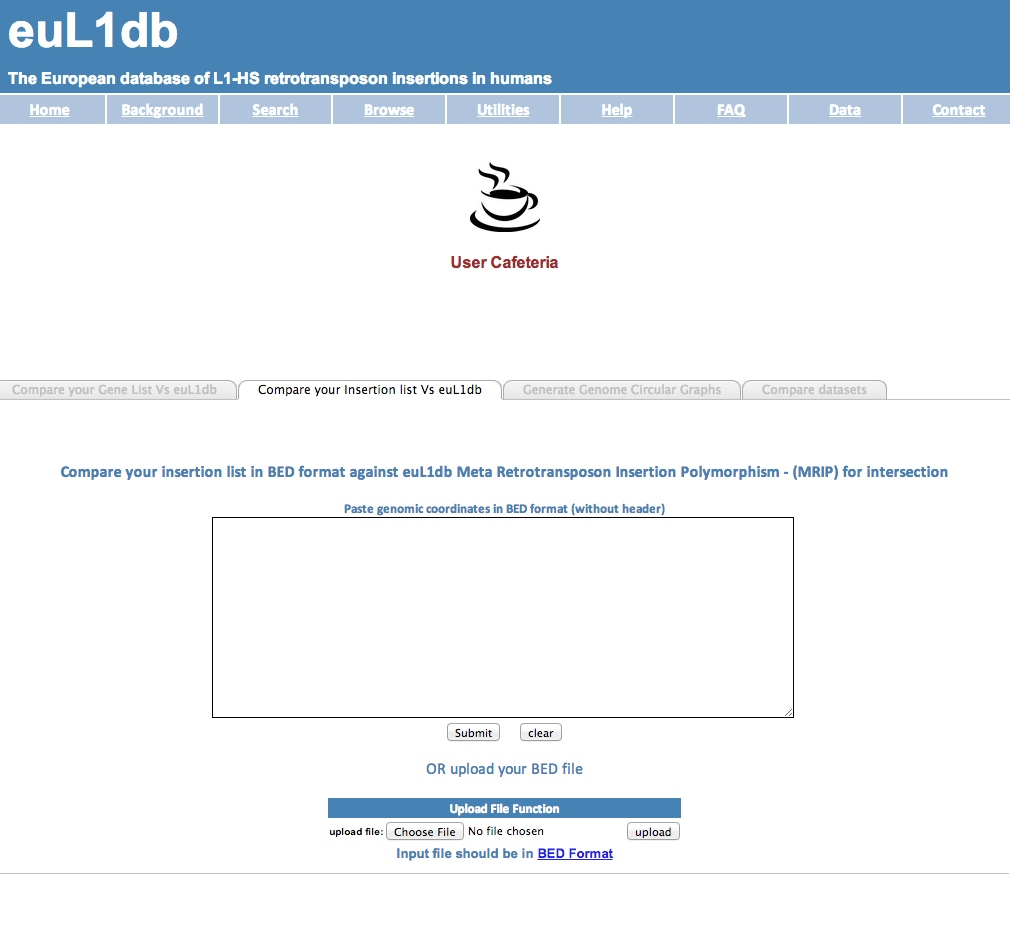
| (B) compare a custom list of L1-HS insertions provided as a list of genomic intervals in BED format with euL1db content. The output is a list of MRIPs intersecting with the genomic intervals provided in the input file; |
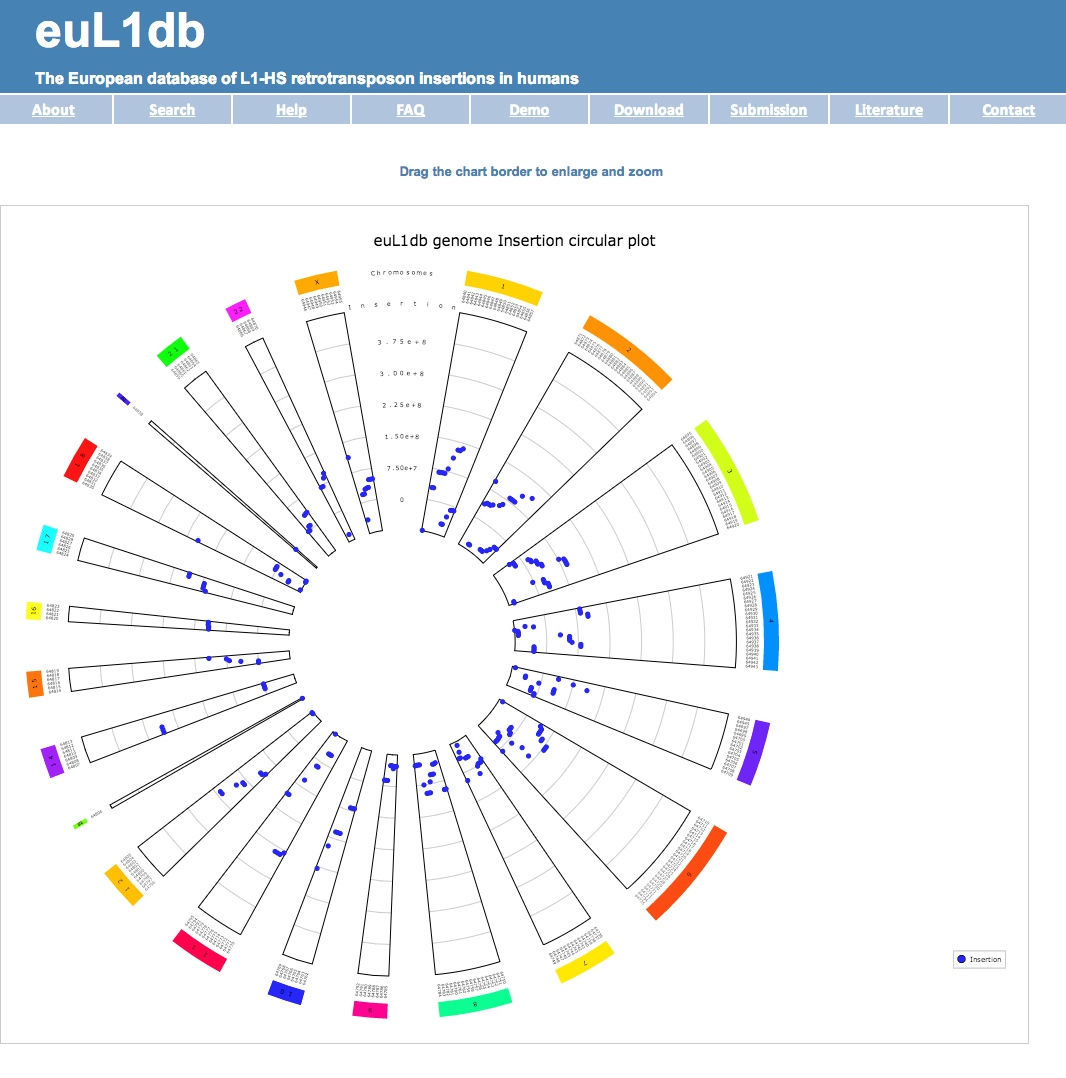
| (C) Display genome circular graphs for a particular study. There is also an option to display insertions per chromosome; |
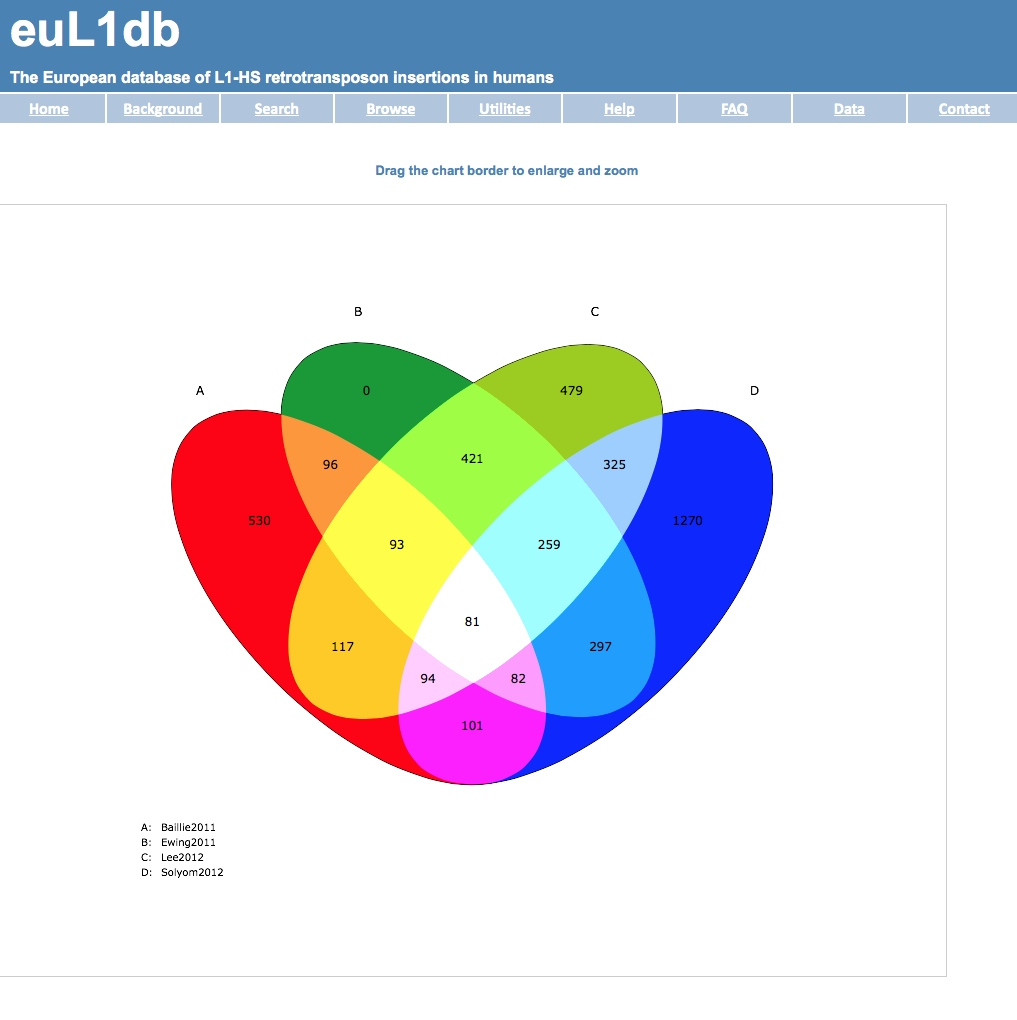
| (D) Compare the datasets present in two or more studies and plot the result as Venn diagrams. AND or NOT operators can be used. |